|
From rxpgnews.com Cytology
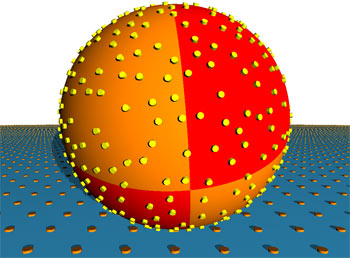
Blood is the universal means with which different types of cells are transported in our bodies. Its movement is determined by hydrodynamic forces. The cells anchor themselves to the walls of the blood vessels in the target tissue with the aid of special adhesive molecules, which are also called receptors. In many cases these receptors are grouped in the cell surface in nanometer-sized patches. The adhesion process is based on the key and lock principle: as a rule, an adhesion molecule only bonds with specific partners. This guarantees that the cells are only brought to a halt where they are to fulfill their biological function.
The initial simulations investigating the influence of flow speed on adhesion revealed that the faster the flow of the liquid, the faster the cells find their adhesion partners as the cell can scan a larger area. The researchers then varied the density of the patches and established that beyond a threshold value of a few hundred receptor areas per cell, there was no further acceleration of adhesion rate because from that point the effective radii of the patches overlap due to their thermal random movement. Similar results were seen with the size of the adhesive areas, which obviously plays a less significant part in effective adhesion. However, changing the height at which the adhesive patches protrude above the cell membrane has surprising results: even small increases give rise to much faster adhesion. White blood corpuscles use this effect by covering themselves with hundreds of protrusions called microvilli, which stand about 350 nanometers above the cell surface - almost four per cent of the cell diametre. Red blood corpuscles infected with malaria also use this "hedgehog spine" strategy. They have "knobs" that are 20 nanometers high on their surface. The scientists suspect that their simulations have helped them to discover a general biological design principle which also occurs in other hydrodynamic contexts - in bacteria, for example, which collect in medical devices through which liquids flow, such as catheters or dialysis equipment. In the future, the software they have developed will allow these situations to be examined more closely than ever before and is another step on the way to "computational" biology. All rights reserved by www.rxpgnews.com |